DNA Sequence Analysis Shows Ebola Outbreak Naturally Ocurring, Not Engineered Virus
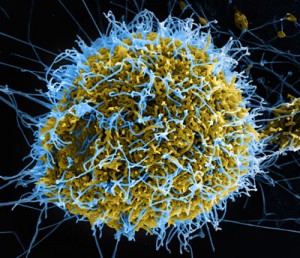
In an electron microscope image that has been colorized, Ebola virus particles in blue are being extruded from an African Green Monkey kidney cell in yellow, grown in a laboratory cell culture system. Photo produced by National Institute of Allergy and Infectious Diseases, NIH.
I had really hoped I wasn’t going to have to write this post. Yesterday, Marcy emailed me a link to a Washington’sBlog post that breathlessly asks us “Was Ebola Accidentally Released from a Bioweapons Lab In West Africa?” Sadly, that post relies on an interview with Francis Boyle, whom I admire greatly for his work as a legal scholar on bioweapons. My copy of his book is very well-thumbed. But Boyle and WashingtonsBlog are just wrong here, and it takes only seconds to prove them wrong.
Shortly after getting the email and reading the blog post, I sent out tweets to this summary and this original scientific report which describe work on DNA analysis of Ebola isolated from multiple patients during the current outbreak. That work conclusively shows that the virus in the current outbreak is intimately related to isolates from previous outbreaks with changes only on the order of the naturally occurring mutation rate known for the virus. Further, these random mutations are spread evenly throughout the short run of the virus’s genes and there are clearly no new bits spliced in by a laboratory. Since I wasn’t seeing a lot of traction from the Washington’sBlog post, I was going to let it just sit there.
I should have alerted last night when I heard my wife chuckling over the line “It is difficult to describe working with a horse infected with Ebola”, but I merely laughed along with her and didn’t ask where she read it.
This morning, while perusing the Washington Post, I saw that Joby Warrick has returned to his beat as the new Judy Miller. Along with the line about the Ebola-infected horse, Warrick’s return to beating the drums over bioweapons fear boasts a headline that could have been penned by WashingtonsBlog: “Ebola crisis rekindles concerns about secret research in Russian military labs“.
Warrick opens with a re-telling of a tragic accident in 1996 in a Soviet lab where a technician accidentally infected herself with Ebola. He uses that to fan flames around Soviet work in that era:
The fatal lab accident and a similar one in 2004 offer a rare glimpse into a 35-year history of Soviet and Russian interest in the Ebola virus. The research began amid intense secrecy with an ambitious effort to assess Ebola’s potential as a biological weapon, and it later included attempts to manipulate the virus’s genetic coding, U.S. officials and researchers say. Those efforts ultimately failed as Soviet scientists stumbled against natural barriers that make Ebola poorly suited for biowarfare.
The bioweapons program officially ended in 1991, but Ebola research continued in Defense Ministry laboratories, where it remains largely invisible despite years of appeals by U.S. officials to allow greater transparency. Now, at a time when the world is grappling with an unprecedented Ebola crisis, the wall of secrecy surrounding the labs looms still larger, arms-control experts say, feeding conspiracy theories and raising suspicions.
/snip/
Enhancing the threat is the facilities’ collection of deadly germs, which presumably includes the strains Soviet scientists tried to manipulate to make them hardier, deadlier and more difficult to detect, said Smithson, now a senior fellow with the James Martin Center for Nonproliferation Studies, a research institute based in Monterey, Calif.
“We have ample accounts from defectors that these are not just strains from nature, but strains that have been deliberately enhanced,” she said.
Only when we get three paragraphs from the end of the article do we get the most important bit of information to be gleaned from the Soviet work on Ebola:
Ultimately, the effort to concoct a more dangerous form of Ebola appears to have failed. Mutated strains died quickly, and Soviet researchers eventually reached a conclusion shared by many U.S. biodefense experts today: Ebola is a poor candidate for either biological warfare or terrorism, compared with viruses such as smallpox, which is highly infectious, or the hardy, easily dispersible bacteria that causes anthrax.
Note also that, in order to make Ebola more scary, Warrick completely fails to mention the escape of weaponized anthrax from a Soviet facility in 1979, infecting 94 and killing 64, dwarfing the toll from the two Ebola accidents.
And lest we calm down about Ebola and the other bioweapons the Soviets worked on, Warrick leaves us this charming tidbit to end the article:
“One must assume that whatever genetically engineered bacterial and viral forms were created . . . remain stored in the culture collections of the Russian Federation Ministry of Defense.”
Okay, so after we finish peeing our pants over the warnings from WashingtonsBlogPost, here are the clear scientific data showing that the virus actually circulating in West Africa fits perfectly within the genetics one would expect from a natural outbreak. From the summary article, we have this:
For their study, published in the August 28 online issue of Science, Gire’s group sequenced viral DNA of samples collected from 78 confirmed Ebola patients in Sierra Leone between late May and mid-June. For 13 of these patients, they collected samples at multiple time points, resulting in a total of 99 viral genome sequences. They compared these Ebola genomes to each other, as well as to three published genomes from Guinea, and 20 sequences generated from previous Ebola outbreaks.
The genomic analysis revealed that the current version of the virus in West Africa most likely spread from Middle Africa within the past 10 years. They also found that the viruses causing this outbreak and the two previous ones diverged from a common ancestor around 2004. This means that these outbreaks arose from different “jumps” from the animal reservoir to the human population. The similarity between samples from the current outbreak confirm that it originated from a single jump, and since that time the disease has spread exclusively from human to human. This is different from previous outbreaks, which had spread via multiple zoonotic events.
If we go to the paper in Science, here are the details of what was found in the DNA sequencing:
We combined the 78 Sierra Leonean sequences with three published Guinean samples (3) [correcting 21 likely sequencing errors in the latter (6)] to obtain a data set of 81 sequences. They reveal 341 fixed substitutions (35 nonsynonymous, 173 synonymous, and 133 noncoding) between the 2014 EBOV and all previously published EBOV sequences, with an additional 55 single-nucleotide polymorphisms (SNPs; 15 nonsynonymous, 25 synonymous, and 15 noncoding), fixed within individual patients, within the West African outbreak. Notably, the Sierra Leonean genomes differ from PCR probes for four separate assays used for EBOV and pan-filovirus diagnostics (table S3).
Deep-sequence coverage allowed identification of 263 iSNVs (73 nonsynonymous, 108 synonymous, 70 noncoding, and 12 frameshift) in the Sierra Leone patients (6). For all patients with multiple time points, consensus sequences were identical and iSNV frequencies remained stable (fig. S4). One notable intrahost variation is the RNA editing site of the glycoprotein (GP) gene (fig. S5A) (10–12), which we characterized in patients (6).
So they found a few hundred single changes in the coding sequence of the virus, spread throughout the genome of the virus. The most important bit of the work is the next paragraph:
Phylogenetic comparison to all 20 genomes from earlier outbreaks suggests that the 2014 West African virus likely spread from central Africa within the past decade. Rooting the phylogeny using divergence from other ebolavirus genomes is problematic (Fig. 2A and fig. S6) (6, 13). However, rooting the tree on the oldest outbreak reveals a strong correlation between sample date and root-to-tip distance, with a substitution rate of 8 × 10−4 per site per year (Fig. 2B and fig. S7) (13). This suggests that the lineages of the three most recent outbreaks all diverged from a common ancestor at roughly the same time, around 2004 (Fig. 2C and Fig. 3A), which supports the hypothesis that each outbreak represents an independent zoonotic event from the same genetically diverse viral population in its natural reservoir.
Translating from the technical language here, what the scientists are saying is that if they compare the DNA sequence data from this outbreak to data from previous outbreaks, it is clear that all of the isolates seen are quite similar. The computer programs for graphically representing these relationships are thrown off slightly by the facts that there is diversity in the pool of viruses circulating in the wild and that the virus also tends to mutate over time. By making the logical assumption of “rooting” the relationships among isolates by putting the oldest one at the bottom of the “tree”, the relationships then all fit perfectly and allow a calculation of the mutation rate over time. Simply put, if the virus circulating now were a product of laboratory manipulation to change the virus, it is very likely that the number of changes that would have been introduced would have blown up the phyologenetic analysis of the current outbreak virus when compared to previous outbreaks. The only way an engineered virus could be involved in this current outbreak would be if somehow a scientist understood how just a very small number of single nucleotide changes could make this virus suddenly more virulent.
While there is a hint that perhaps this virus may be more virulent in the evidence that this outbreak may trace to only one jump from a host species instead of several (although I’ve seen analyses suggesting that this outbreak was just unlucky in getting to highly populated areas quickly, accounting for its spread) the authors of the study demonstrate that our knowledge of Ebola is not yet at a level where one could put just those few changes into the genome to achieve higher virulence. In fact, one of the driving reasons for carrying out this study was to identify just those changes that can affect virulence so that the information can possibly be put to use in developing vaccines or other treatments, as seen in the final two paragraphs of the publication:
As in every EVD outbreak, the 2014 EBOV variant carries a number of genetic changes distinct to this lineage; our data do not address whether these differences are related to the severity of the outbreak. However, the catalog of 395 mutations, including 50 fixed nonsynonymous changes with 8 at positions with high levels of conservation across ebolaviruses, provides a starting point for such studies (table S4).
To aid in relief efforts and facilitate rapid global research, we have immediately released all sequence data as it is generated. Ongoing epidemiological and genomic surveillance is imperative to identify viral determinants of transmission dynamics, monitor viral changes and adaptation, ensure accurate diagnosis, guide research on therapeutic targets, and refine public health strategies. It is our hope that this work will aid the multidisciplinary international efforts to understand and contain this expanding epidemic.
As a sad postscript, the paper is dedicated to five health care workers among the paper’s authors who died of Ebola during the time the manuscript was in preparation:
Tragically, five co-authors, who contributed greatly to public health and research efforts in Sierra Leone, contracted EVD and lost their battle with the disease before this manuscript could be published: Mohamed Fullah, Mbalu Fonnie, Alex Moigboi, Alice Kovoma, and S. Humarr Khan. We wish to honor their memory.